728x90
반응형
아래 출처를 통해, Data 분석 및 XGBoost 진행하였음
아직 이해하지 못하거나, 개념적인 부분이 미흡함
개념 부분을 공부해서 정리할 예정
## MRI and Alzheimers
# 데이터출처 : https://www.kaggle.com/datasets/jboysen/mri-and-alzheimers
In [2]:
import tensorflow.compat.v1 as tf
from sklearn.metrics import confusion_matrix
import numpy as np
from scipy.io import loadmat
import os
from pywt import wavedec
from functools import reduce
from scipy import signal
from scipy.stats import entropy
from scipy.fft import fft, ifft
import pandas as pd
from sklearn.model_selection import train_test_split, StratifiedKFold
from sklearn.model_selection import RandomizedSearchCV
from sklearn.preprocessing import StandardScaler
from tensorflow import keras as K
import matplotlib.pyplot as plt
import scipy
from sklearn import metrics
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score
from sklearn.model_selection import KFold,cross_validate
from tensorflow.keras.layers import Dense, Activation, Flatten, concatenate, Input, Dropout, LSTM, Bidirectional,BatchNormalization,PReLU,ReLU,Reshape
from sklearn.metrics import classification_report
from tensorflow.keras.models import Sequential, Model, load_model
import matplotlib.pyplot as plt;
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
from sklearn.decomposition import PCA
from sklearn.model_selection import cross_val_score
from tensorflow.keras.layers import Conv1D,Conv2D,Add
from tensorflow.keras.layers import MaxPool1D, MaxPooling2D
import seaborn as sns
In [3]:
# os.environ을 이용하여 Kaggle API Username, Key 세팅하기
os.environ['KAGGLE_USERNAME'] = ''
os.environ['KAGGLE_KEY'] = ''
In [4]:
# Linux 명령어로 Kaggle API를 이용하여 데이터셋 다운로드하기 (!kaggle ~)
# Linux 명령어로 압축 해제하기
!kaggle datasets download -d jboysen/mri-and-alzheimers
!unzip '*.zip'
mri-and-alzheimers.zip: Skipping, found more recently modified local copy (use --force to force download)
Archive: mri-and-alzheimers.zip
replace oasis_cross-sectional.csv? [y]es, [n]o, [A]ll, [N]one, [r]ename: In [13]:
!ls
mri-and-alzheimers.zip oasis_longitudinal.csv
oasis_cross-sectional.csv sample_data
In [14]:
# pd.read_csv()로 csv파일 읽어들이기
data_cross = pd.read_csv("oasis_cross-sectional.csv")
data_long = pd.read_csv("oasis_longitudinal.csv")
print(data_cross.info())
print(data_long.info())
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 436 entries, 0 to 435
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ID 436 non-null object
1 M/F 436 non-null object
2 Hand 436 non-null object
3 Age 436 non-null int64
4 Educ 235 non-null float64
5 SES 216 non-null float64
6 MMSE 235 non-null float64
7 CDR 235 non-null float64
8 eTIV 436 non-null int64
9 nWBV 436 non-null float64
10 ASF 436 non-null float64
11 Delay 20 non-null float64
dtypes: float64(7), int64(2), object(3)
memory usage: 41.0+ KB
None
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 373 entries, 0 to 372
Data columns (total 15 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Subject ID 373 non-null object
1 MRI ID 373 non-null object
2 Group 373 non-null object
3 Visit 373 non-null int64
4 MR Delay 373 non-null int64
5 M/F 373 non-null object
6 Hand 373 non-null object
7 Age 373 non-null int64
8 EDUC 373 non-null int64
9 SES 354 non-null float64
10 MMSE 371 non-null float64
11 CDR 373 non-null float64
12 eTIV 373 non-null int64
13 nWBV 373 non-null float64
14 ASF 373 non-null float64
dtypes: float64(5), int64(5), object(5)
memory usage: 43.8+ KB
None
In [15]:
print(data_cross.isna().sum())
print("\n")
data_long.isna().sum()
ID 0
M/F 0
Hand 0
Age 0
Educ 201
SES 220
MMSE 201
CDR 201
eTIV 0
nWBV 0
ASF 0
Delay 416
dtype: int64
Out[15]:
Subject ID 0
MRI ID 0
Group 0
Visit 0
MR Delay 0
M/F 0
Hand 0
Age 0
EDUC 0
SES 19
MMSE 2
CDR 0
eTIV 0
nWBV 0
ASF 0
dtype: int64In [16]:
data_cross.dropna(subset=['CDR'],inplace=True)
In [17]:
data_cross.drop(columns=['ID','Delay'],inplace=True)
data_long = data_long.rename(columns={'EDUC':'Educ'})
data_long.drop(columns=['Subject ID','MRI ID','Group','Visit','MR Delay'],inplace=True)
In [18]:
data = pd.concat([data_cross,data_long], ignore_index = True)
data.head()
Out[18]:
M/FHandAgeEducSESMMSECDReTIVnWBVASF01234
| F | R | 74 | 2.0 | 3.0 | 29.0 | 0.0 | 1344 | 0.743 | 1.306 |
| F | R | 55 | 4.0 | 1.0 | 29.0 | 0.0 | 1147 | 0.810 | 1.531 |
| F | R | 73 | 4.0 | 3.0 | 27.0 | 0.5 | 1454 | 0.708 | 1.207 |
| M | R | 74 | 5.0 | 2.0 | 30.0 | 0.0 | 1636 | 0.689 | 1.073 |
| F | R | 52 | 3.0 | 2.0 | 30.0 | 0.0 | 1321 | 0.827 | 1.329 |
In [19]:
data.describe()
Out[19]:
AgeEducSESMMSECDReTIVnWBVASFcountmeanstdmin25%50%75%max
| 608.000000 | 608.000000 | 570.00000 | 606.000000 | 608.000000 | 608.000000 | 608.00000 | 608.000000 |
| 75.208882 | 10.184211 | 2.47193 | 27.234323 | 0.288651 | 1477.062500 | 0.73713 | 1.203597 |
| 9.865026 | 6.058388 | 1.12805 | 3.687980 | 0.377697 | 170.653795 | 0.04267 | 0.135091 |
| 33.000000 | 1.000000 | 1.00000 | 4.000000 | 0.000000 | 1106.000000 | 0.64400 | 0.876000 |
| 70.000000 | 4.000000 | 2.00000 | 26.000000 | 0.000000 | 1352.500000 | 0.70400 | 1.118000 |
| 76.000000 | 12.000000 | 2.00000 | 29.000000 | 0.000000 | 1460.000000 | 0.73600 | 1.202000 |
| 82.000000 | 16.000000 | 3.00000 | 30.000000 | 0.500000 | 1569.000000 | 0.76625 | 1.297500 |
| 98.000000 | 23.000000 | 5.00000 | 30.000000 | 2.000000 | 2004.000000 | 0.84700 | 1.587000 |
In [20]:
cor = data.corr()
plt.figure(figsize=(12,9))
sns.heatmap(cor, xticklabels=cor.columns.values,yticklabels=cor.columns.values, annot=True)
Out[20]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f6c54220710>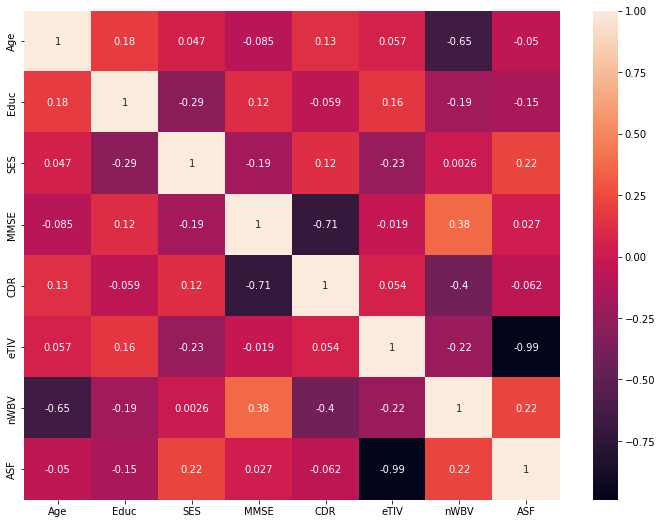
In [29]:
data.isna().sum()
Out[29]:
M/F 0
Hand 0
Age 0
Educ 0
SES 38
MMSE 2
CDR 0
eTIV 0
nWBV 0
ASF 0
dtype: int64In [30]:
from sklearn.impute import SimpleImputer
imputer = SimpleImputer ( missing_values = np.nan,strategy='most_frequent')
imputer.fit(data[['SES']])
data[['SES']] = imputer.fit_transform(data[['SES']])
# We perform it with the median
imputer = SimpleImputer ( missing_values = np.nan,strategy='median')
imputer.fit(data[['MMSE']])
data[['MMSE']] = imputer.fit_transform(data[['MMSE']])
In [31]:
data
Out[31]:
M/FHandAgeEducSESMMSECDReTIVnWBVASF01234...603604605606607
| F | R | 74 | 2.0 | 3.0 | 29.0 | 0.0 | 1344 | 0.743 | 1.306 |
| F | R | 55 | 4.0 | 1.0 | 29.0 | 0.0 | 1147 | 0.810 | 1.531 |
| F | R | 73 | 4.0 | 3.0 | 27.0 | 0.5 | 1454 | 0.708 | 1.207 |
| M | R | 74 | 5.0 | 2.0 | 30.0 | 0.0 | 1636 | 0.689 | 1.073 |
| F | R | 52 | 3.0 | 2.0 | 30.0 | 0.0 | 1321 | 0.827 | 1.329 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| M | R | 82 | 16.0 | 1.0 | 28.0 | 0.5 | 1693 | 0.694 | 1.037 |
| M | R | 86 | 16.0 | 1.0 | 26.0 | 0.5 | 1688 | 0.675 | 1.040 |
| F | R | 61 | 13.0 | 2.0 | 30.0 | 0.0 | 1319 | 0.801 | 1.331 |
| F | R | 63 | 13.0 | 2.0 | 30.0 | 0.0 | 1327 | 0.796 | 1.323 |
| F | R | 65 | 13.0 | 2.0 | 30.0 | 0.0 | 1333 | 0.801 | 1.317 |
608 rows × 10 columns
In [33]:
from sklearn import preprocessing
le = preprocessing.LabelEncoder()
data['CDR'] = le.fit_transform(data['CDR'].values)
In [34]:
data = pd.get_dummies(data)
In [35]:
plt.bar(data['CDR'].unique(),data['CDR'].value_counts())
Out[35]:
<BarContainer object of 4 artists>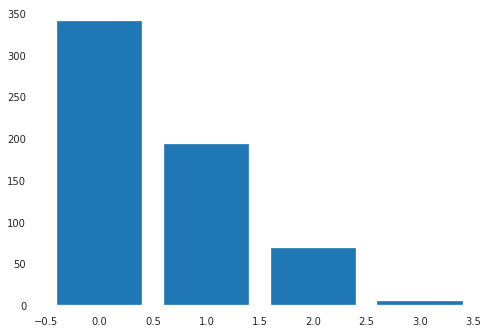
In [36]:
data = data.drop(data[data['CDR']==3].index)
data.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 603 entries, 0 to 607
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Age 603 non-null int64
1 Educ 603 non-null float64
2 SES 603 non-null float64
3 MMSE 603 non-null float64
4 CDR 603 non-null int64
5 eTIV 603 non-null int64
6 nWBV 603 non-null float64
7 ASF 603 non-null float64
8 M/F_F 603 non-null uint8
9 M/F_M 603 non-null uint8
10 Hand_R 603 non-null uint8
dtypes: float64(5), int64(3), uint8(3)
memory usage: 44.2 KB
In [37]:
y = data.pop('CDR')
x = data
In [38]:
x = StandardScaler().fit_transform(x)
In [39]:
X_train, X_test, y_train, y_test = train_test_split(x, y, random_state = 42,test_size=0.3)
In [40]:
from xgboost import XGBClassifier
FOLDS = 5
parametros_xgb = {
"learning_rate": [0.01, 0.025, 0.005,0.5, 0.075, 0.1, 0.15, 0.2,0.3,0.8,0.9],
"max_depth":[3,5,8,10,15,20,25,30,40,50],
"n_estimators":range(1,1000)
}
model_xgb= XGBClassifier(eval_metric='mlogloss')
xgb_random = RandomizedSearchCV(estimator = model_xgb, param_distributions = parametros_xgb, n_iter = 100, cv = FOLDS,
verbose=0, random_state=42,n_jobs = -1, scoring='accuracy')
xgb_random.fit(X_train, y_train)
xgb_random.best_params_
Out[40]:
{'learning_rate': 0.5, 'max_depth': 30, 'n_estimators': 103}In [41]:
model_xgb = xgb_random.best_estimator_
model_xgb.fit(X_train,y_train)
model_xgb.score(X_test,y_test)
Out[41]:
0.7845303867403315In [43]:
from sklearn.ensemble import GradientBoostingClassifier
FOLDS =10
parametros_gb = {
"loss":["deviance"],
"learning_rate": [0.01, 0.025, 0.005,0.5, 0.075, 0.1, 0.15, 0.2,0.3,0.8,0.9],
"min_samples_split": [0.01, 0.025, 0.005,0.4,0.5, 0.075, 0.1, 0.15, 0.2,0.3,0.8,0.9],
"min_samples_leaf": [1,2,3,5,8,10,15,20,40,50,55,60,65,70,80,85,90,100],
"max_depth":[3,5,8,10,15,20,25,30,40,50],
"max_features":["log2","sqrt"],
"criterion": ["friedman_mse", "mae"],
"subsample":[0.5, 0.618, 0.8, 0.85, 0.9, 0.95, 1.0],
"n_estimators":range(1,100)
}
model_gb= GradientBoostingClassifier()
gb_random = RandomizedSearchCV(estimator = model_gb, param_distributions = parametros_gb, n_iter = 100, cv = FOLDS,
verbose=0, random_state=42,n_jobs = -1, scoring='accuracy')
gb_random.fit(X_train, y_train)
gb_random.best_params_
Out[43]:
{'criterion': 'mae',
'learning_rate': 0.8,
'loss': 'deviance',
'max_depth': 20,
'max_features': 'log2',
'min_samples_leaf': 1,
'min_samples_split': 0.025,
'n_estimators': 68,
'subsample': 0.95}In [44]:
model_gb = gb_random.best_estimator_
print(model_gb.score(X_test,y_test))
0.8232044198895028
In [45]:
cross_val_score(model_gb, x, y, cv=10, scoring='accuracy').mean()
Out[45]:
0.7856284153005465In [46]:
y_pred = model_gb.predict(X_test)
cm = confusion_matrix(y_test, y_pred)
clr = classification_report(y_test, y_pred)
plt.figure(figsize=(8, 8))
sns.heatmap(cm, annot=True, vmin=0, fmt='g', cbar=False, cmap='Blues')
plt.xlabel("Predicted")
plt.ylabel("Actual")
plt.title("Confusion Matrix")
plt.show()
print("Classification Report:\n----------------------\n", clr)
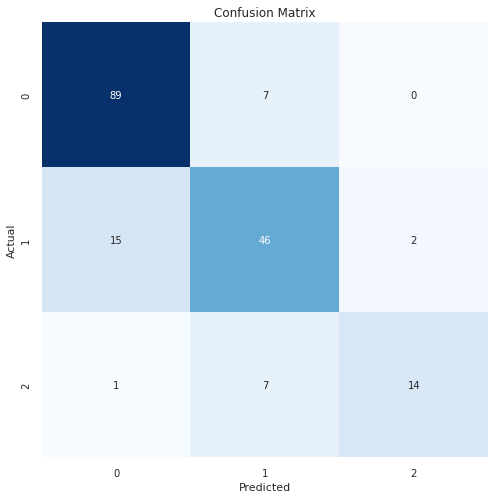
Classification Report:
----------------------
precision recall f1-score support
0 0.85 0.93 0.89 96
1 0.77 0.73 0.75 63
2 0.88 0.64 0.74 22
accuracy 0.82 181
macro avg 0.83 0.76 0.79 181
weighted avg 0.82 0.82 0.82 181
728x90
반응형
'Data Analysis > Machine Learning' 카테고리의 다른 글
| Seaborn을 활용한 데이터 분포 시각화 17가지 방법 (1) | 2022.05.08 |
|---|---|
| train_test_split 학습데이터와 테스트데이터 분리 (0) | 2022.05.06 |
| StandardScaler를 이용하여 데이터 전처리 (0) | 2022.05.05 |
| XGBoost (0) | 2022.05.05 |
댓글